Introduction
We have upgraded our previous web server CPGAVAS into CPGAVAS2 (an integrated Plastome Annotator and Analyzer), with the addition of new functions.
- We constructed two datasets. The 43-plastome dataset is curated with RNA-seq data, and the 2544-plastome dataset contains the largest number of plastome sequences among those similar plastome sequence annotation tools. In addition, CPGAVAS2 accepts user-provided reference sequence.
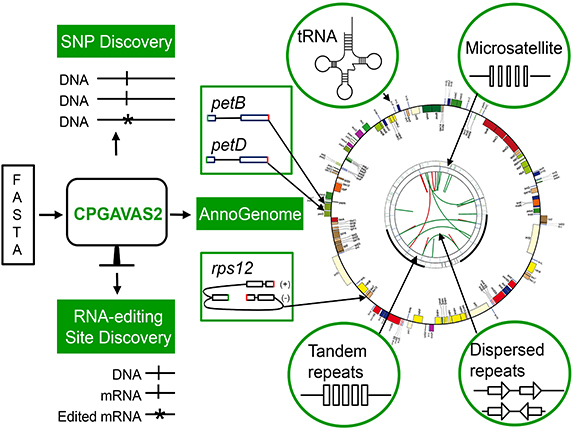
- In addition to predicting structurally simple genes, we developed two algorithms to annotate structurally complex genes such as those having small exons (petB, petD and rpl16) or trans-splicing exons (rps12).
- CPGAVAS2 discovers repeats automatically after annotating the genome, whose results are presented in a circular map together with the annotation of genes.
- CPGAVAS2 supports the exploratory analyses of plastome diversity by identifying SNPs and RNA editing sites if user supplies the Next Generation Sequencing (NGS) data.
The command line version of CPGAVAS2 has been released. Please click here for more information.
We believe CPGAVAS2 will become a powerful tool for plastome research in the NGS era.
The Main Modules of CPGAVAS2
- "AnnotateGenome"- This module can annotate a plastome sequence when user provides a plastome sequence in FASTA format and optional annotation file in GenBank format.
- "ViewResults"- This module allows the retrieval and examination of analysis results from all modules.
- "UpdateAnnotatioinResults"- The manually curated gene annotation information in GFF3 format file can be re-analyzed using this function. It will reproduce the circular map and the analysis results.
- "ExtractSeq"- This module allows a user to retrieve the sequences for a list of plastid genes for a list of species. These data will be used for phylogenomic analyses.
- "AnaDiversity"- This module supports the preliminary identification of Single-nucleotide polymorphisms (SNP) and the prediction of RNA editing sites using NGS data. However, the results will depend on the setup of particular experiments.
The Overall Workflow of CPGAVAS2
The input for CPGAVAS2 is a plastome sequence and the output includes the gene models in GFF3 format, circular map in PNG format, analysis results and files for GenBank submission. A workflow is shown below